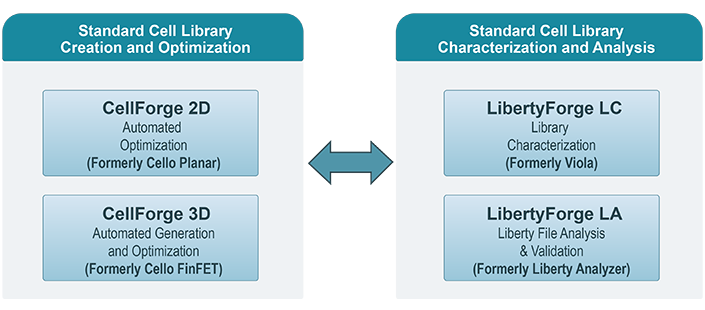
Automated Library Creation, Optimization and Characterization
Silvaco’s Automated Standard Cell Library creation, optimization, and characterization solutions deliver the most comprehensive tool suite for development of standard cell libraries. Our solutions enable standard cell library designers to deliver complete libraries, fully characterized and validated, in weeks instead of months enabling rapid time to market and development cost reduction.
Library Optimization
Library Characterization and Variation Modeling
Advanced Technology Node Layout Generation
Liberty Analyzer for Liberty Files
Automated Optimization
CellForge 2D™ is the industry’s most versatile, integrated and easy-to-use solution for digital cell library optimization. It enables digital CMOS IC designers to explore the impact of alternate device models, design rules and cell architectures. The underlying framework allows designers to centralize their library development into a single location and provides a rich interface for customization.
Advanced Technology Node Layout Generation
CellForge 3D™ offers a cutting-edge solution for advanced technology nodes, leveraging their strict design constraints to employ powerful algorithms that automate the layout generation from transistor netlists. This solution allows digital IC designers to explore and encode constrains to optimize power, performance, and area (PPA) metrics of their libraries, evaluate the impact of design-technology co-optimization (DTCO) strategies, and assess emerging innovations early in the design phase.
Library Characterization
LibertyForge LC™ is a unique, all-inclusive package for fast and accurate characterization of digital cell libraries. The package includes a powerful SPICE-based characterization engine with fully automated stimulus generation, a library model checker and a databook generator.
Liberty File Validation
LibertyForge LA™ displays, analyzes, compares, and validates Liberty™ files for timing, power, noise, and area. LibertyForge LA handles multiple NLDM, NLPM, CCS, ECSM, and LVF models at library, cell, pin, and individual arc levels while providing insightful statistical data.